Table of Contents
1. What is bacterial aromatic polyketide?
Polyketides are a large family of secondary metabolites with diverse structures and biological activities. Many of them are clinically important compounds such as anti-biotic, anti-fungal, and anti-cancer drugs [1]. The biosynthesis of polyketide is catalyzed by a collection of enzymes called polyketide synthases (PKSs). The carbon chain of polyketide is formed through the stepwise decarboxylative condensation of acyl-thioester units using coordinated group of PKS domains. The genes encoding PKS are usually clustered with their auxiliary and regulatory elements on genome and their products are classified into type I, II, and III depending on the organization of their domains [2].
 Das A, Khosla C.: Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009, 42:631-9.
Das A, Khosla C.: Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009, 42:631-9.
Bacterial aromatic polyketides such as tetracyclines and actinorhodin are polycyclic phenolic compounds that are assembled by type II PKSs with at most two domains. A hallmark of these multi-enzyme complexes is the iterative use of a ketosynthase heterodimer (KS and CLF) that catalyze Claisen condensations of theoesters with malonyl-CoA units [3]. During this process, acyl carrier protein (ACP) delivers malonyl building blocks to the ketosynthase heterodimer. Besides these minimal PKSs components, additional tailoring enzymes such as ketoreductases (KRs), aromatases (AROs), and cyclises (CYCs) are required to modify the highly reactive poly-beta-keto intermediates to specific folding pattern. Typical primary products of type II PKS from the concerted type II PKS actions are polyphenols in seven polyketide chemotypes such as the linear tetracyclines, anthracyclines, benzoisochromanequinones, tetracenomycins, aureolic acids, and the angular angucyclines as well as a group of pentagular polyphenols [4]. Additional modification by numerous elaborate tailoring enzymes such as dimerases, P450 monooxygenases, methyltransferases, and glycosyltransferases can take place to further diversify the phenolic polycycles [5].
[1] Staunton J, Weissman KJ: Polyketide biosynthesis: a millennium review. Nat Prod Rep 2000, 18: 380-416.
[2] Shen B: Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol 2003, 7:285-95
[3] Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A: Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep 2007, 24:162-90
[4] Fritzsche K, Ishida K, Hertweck C: Orchestration of discoid polyketide cyclization in the resistomycin pathway. J Am Chem Soc 2008, 130:8307-16
[5] Rix U, Fischer C, Remsing LL, Rohr J: Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat Prod Rep 2002, 19:542-80
[1] Staunton J, Weissman KJ: Polyketide biosynthesis: a millennium review. Nat Prod Rep 2000, 18: 380-416.
[2] Shen B: Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol 2003, 7:285-95
[3] Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A: Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep 2007, 24:162-90
[4] Fritzsche K, Ishida K, Hertweck C: Orchestration of discoid polyketide cyclization in the resistomycin pathway. J Am Chem Soc 2008, 130:8307-16
[5] Rix U, Fischer C, Remsing LL, Rohr J: Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat Prod Rep 2002, 19:542-80
2. Biosynthetic function of type II PKS
2.1. Result of known type II PKS using homology based clustering
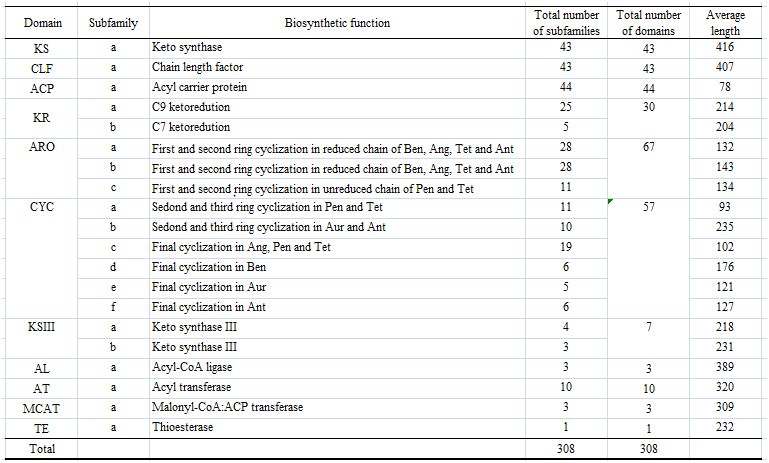
Abbreviation: Unc-unclassified, Ang-Angucyclines, Ant-Anthracyclines, Ben-Benzoisochromanequinones, Pen-Pentangular polyphenols, Tet-Tetracenomycins, Aur-Tetracyclines/aureolic acids
PKMiner includes 280 known type II PKSs identified from 42 known type II PKS gene clusters. For each of type II PKS domain, the above table shows the subfamily, biosynthetic function, total number of domains, total number of subfamilies and the average length present in 280 known type II PKSs
2.2. Map between type II PKS and aromatic polyketide chemotype
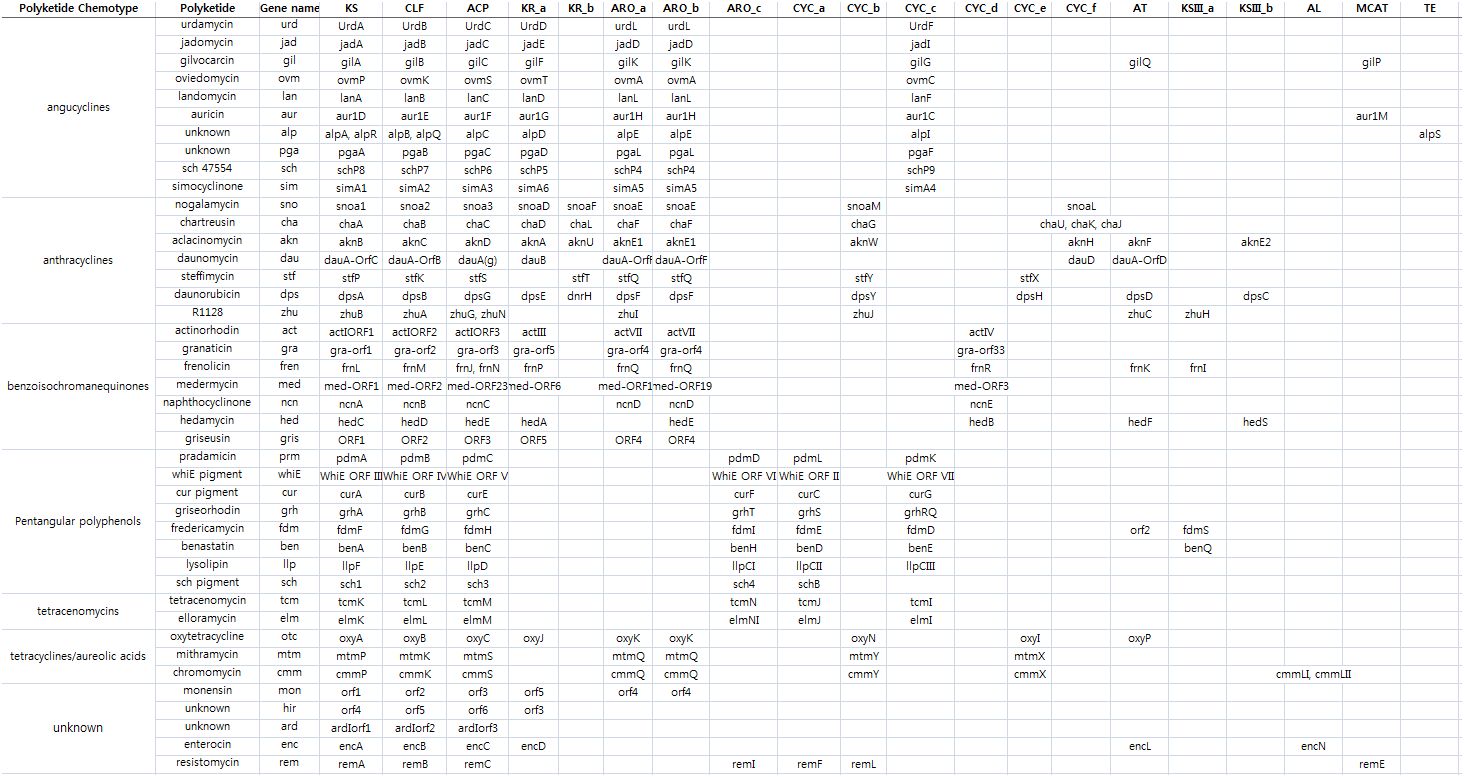
Table shows II PKS domains in each type II PKS gene cluster for each aromatic polyketide chemotypes
3. Type II PKS domain classifiers
3.1. Performance evaluation of type II PKS domain classifiers.
Evaluation of type II PKS domain classifiers using profiled HMM and sequence pairwise alignment based SVM with 4- fold cross-validation (n>20) and leave-one-out cross-validation (n<20)
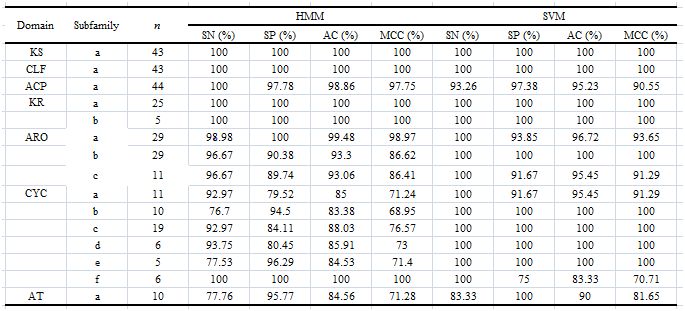
For each type II PKS domain, PKMiner include type II PKS domain classifier with superior classification accuracy
4. Aromatic polyketide chemotype prediction rules
4.1. Type II PKS ARO/CYC domain combinations of aromatic polyketide chemotype
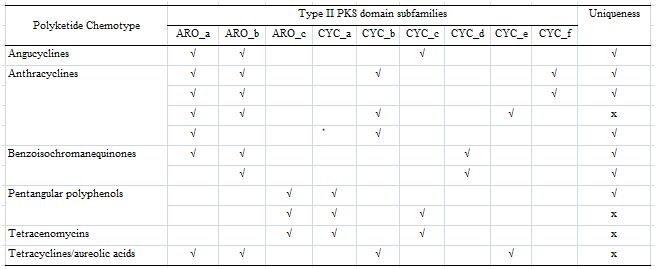
For each aromatic polyketide chemotype, this table shows ARO/CYC domain combinations of type II PKS gene clusters. The uniqueness column indicates whether or not type II PKS ARO/CYC domain combinations overlap between aromatic polyketide chemotypes
4.2. Derivation of prediction rules for aromatic polyketide chemotype
The aromatic polyketide chemotype classification rules based on domain combination and sequence homology are as followings:
For type II PKS gene cluster mapped onto aromatic polyketide chemotype with unique domain combination, we assigned corresponding polyketide chemotype into type II PKS gene cluster.
For type II PKS gene cluster mapped onto aromatic polyketide chemotype with overlapped domain combination, we assigned the most abundant polyketide chemotype of homologs of type II PKS ARO and CYC onto type II PKS gene cluster.
5. PKSDB
5.1. List of polyketide and genome
PKMiner provides known type II PKSs identified from aromatic polyketide gene cluster and predicted type II PKSs resulted from genome analysis.
User can explore the detail information of aromatic polyketide, type II PKS and genome analysis result by clicking the button in detail column
Currently analyzed genome list shows show the total number of genomes for identifying type II PKSs.
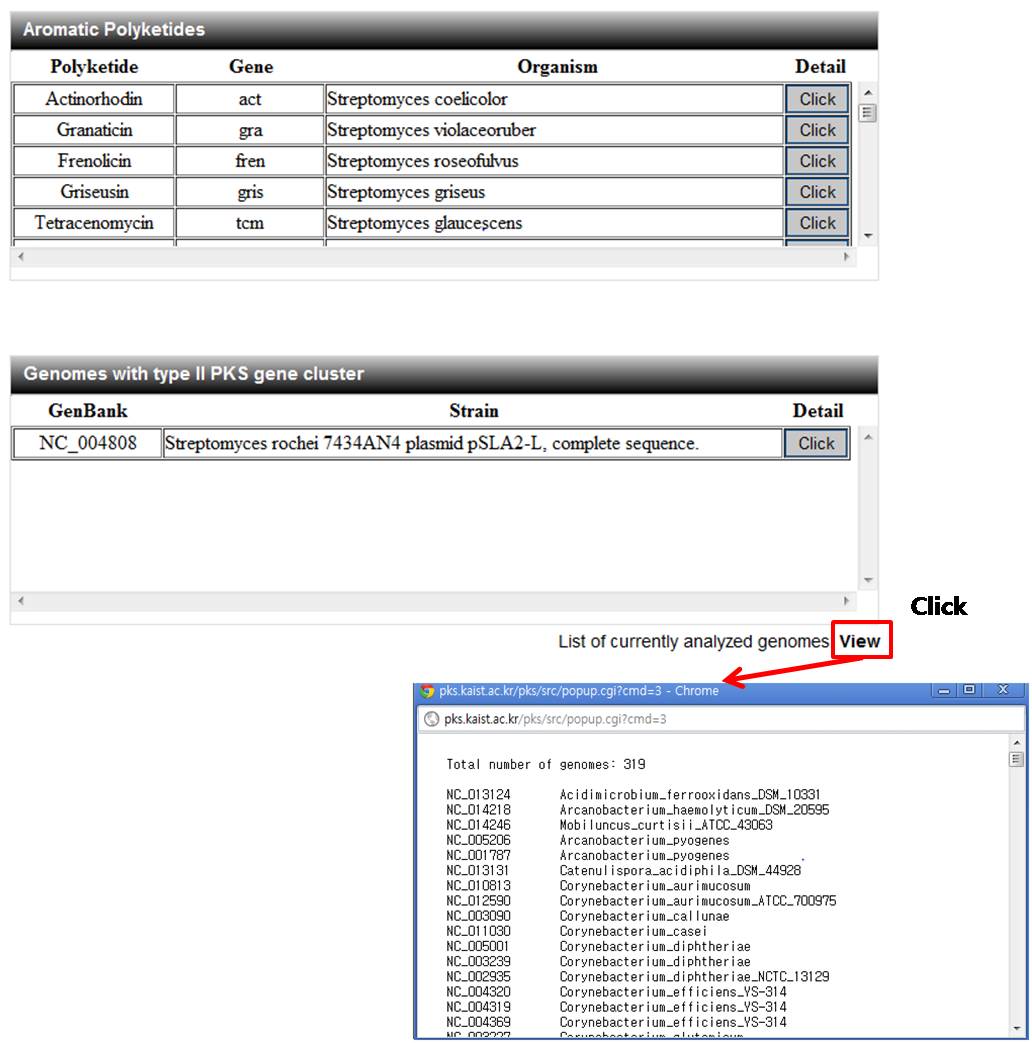
5.2. Aromatic polyketide and their type II PKS information
It provide detail information of aromatic polyketide and their type II PKSs.
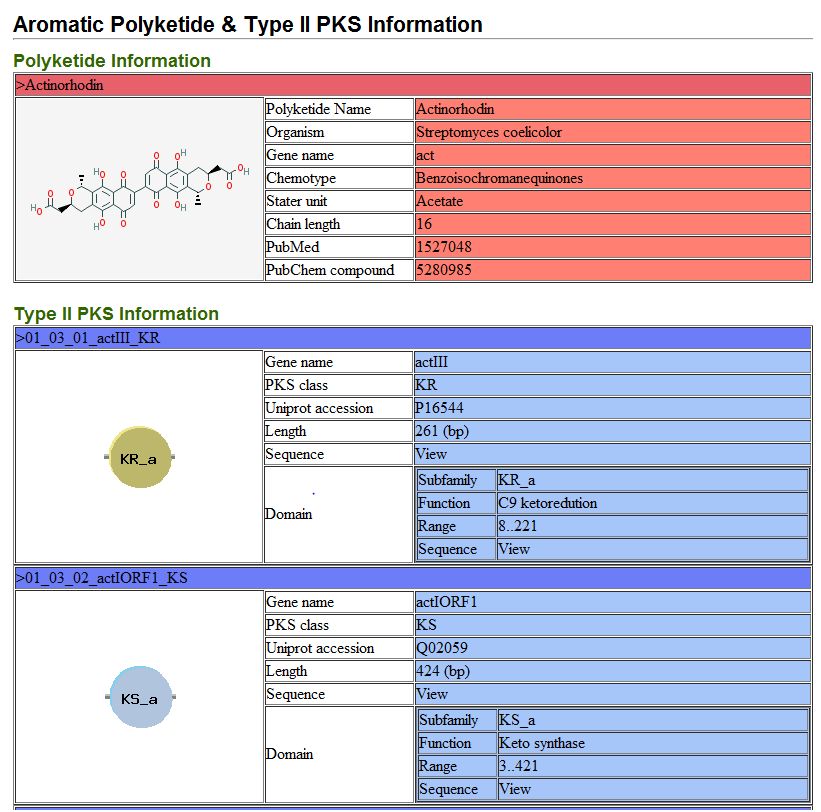
5.3. Analyzed genome information
It provide detail information of type II PKS gene cluster and their type II PKSs from genome analysis result
6. SEARCH
6.1. Sequence input interface
A user finds similar type II PKS to the query using type II PKS domain classifiers. It provides two sequence search option: Evalue and Coverage.
Coverage means that the percentage of query sequence alignment to target sequence.
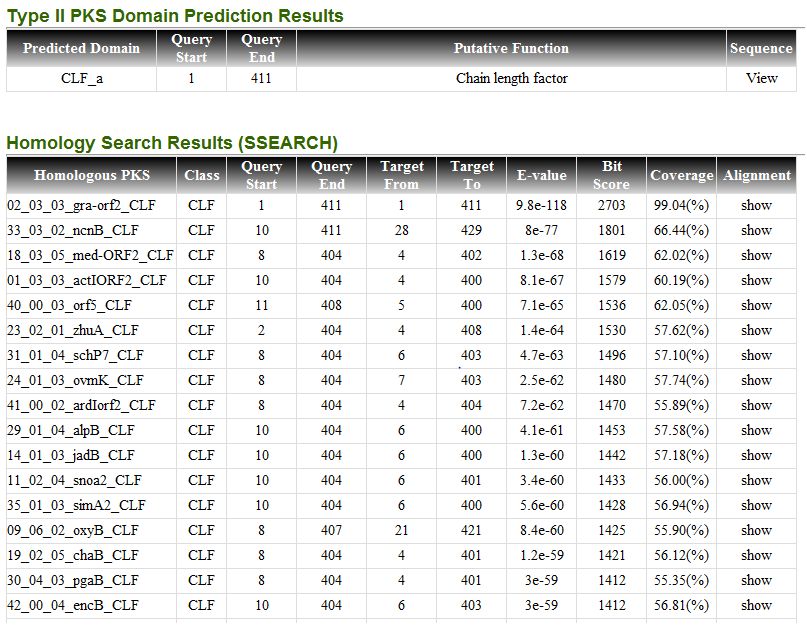
6.2. Sequence search result
Sequence search result include sequence analysis result using type II PKS domain classifiers and sequence homology using ssearch36.
Each entry in homologous PKS is linked to detail information page of related polyketide.
7. GENOME MINING
7.1. Sequence input interface
Genome mining interface provides two methods for genome analysis. User can upload genome sequence in form of genbank or fasta format. User can also insert genbank accession instead of uploading genome sequence.
In case of genome sequence in form of fasta format, PKMiner predict ORF from genome sequence using glimmer trained with genome sequence of Steptomyces Coelicolor.
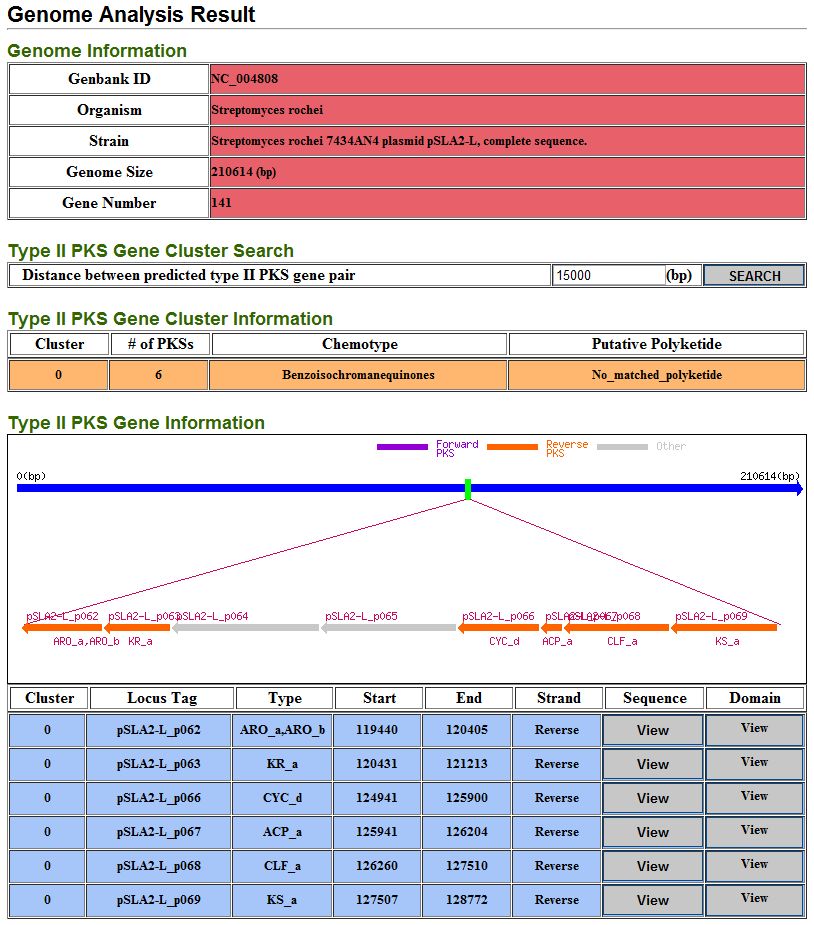
7.2. Genome analysis result
It provide detail information of type II PKS gene cluster and their type II PKSs from genome analysis result.
User can reorganize predicted type II PKSs based on distance between predicted type II PKS gene pair.
It can help identify type II PKS gene cluster.
Copyright 2012 © Bisyn Lab. All Rights Reserved.